Bis-Heteroaryl Pyrazoles: Identification of Orally Bioavailable Inhibitors of Activin Receptor-Like Kinase-2 (R206H).
Sekimata, K., Sato, T., Sakai, N., Watanabe, H., Mishima-Tsumagari, C., Taguri, T., Matsumoto, T., Fujii, Y., Handa, N., Honma, T., Tanaka, A., Shirouzu, M., Yokoyama, S., Miyazono, K., Hashizume, Y., Koyama, H.(2019) Chem Pharm Bull (Tokyo) 67: 224-235
- PubMed: 30828000
- DOI: https://doi.org/10.1248/cpb.c18-00598
- Primary Citation of Related Structures:
6ACR - PubMed Abstract:
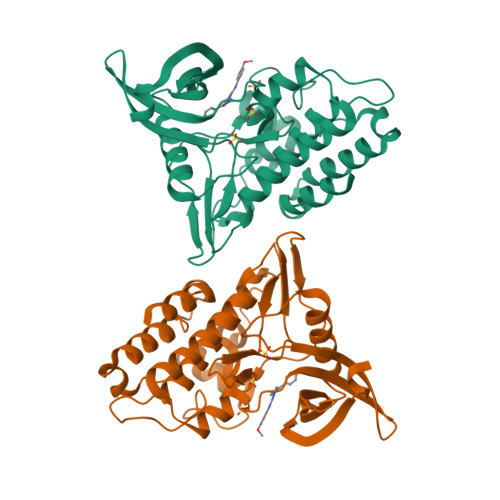
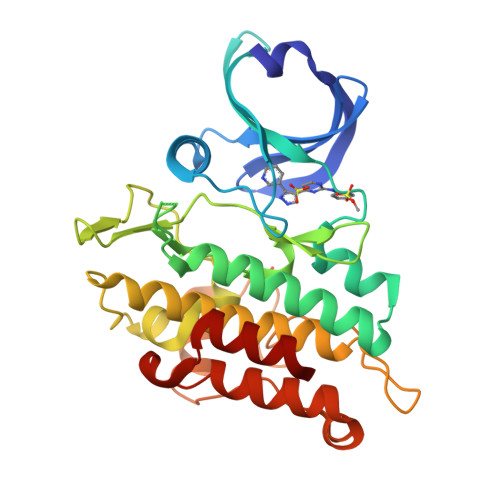
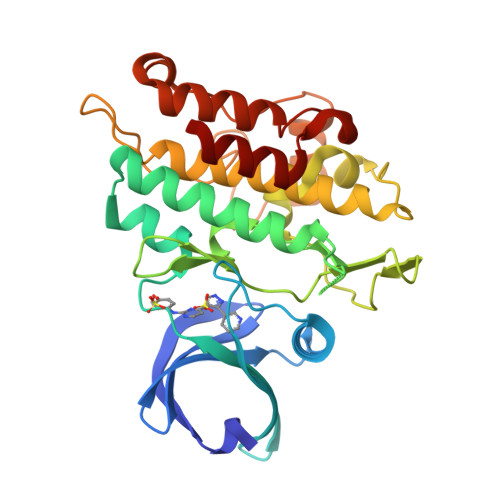
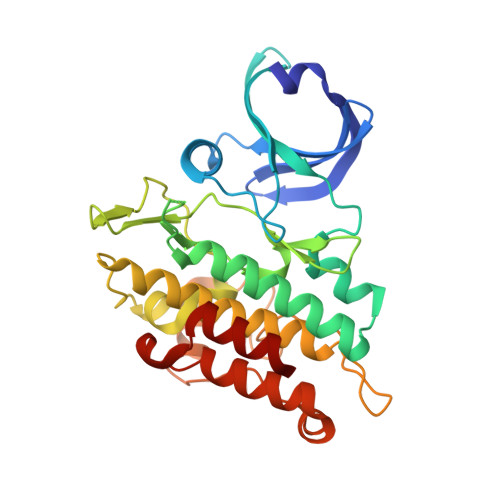
Mutant activin receptor-like kinase-2 (ALK2) was reported to be closely associated with the pathogenesis of fibrodysplasia ossificans progressiva (FOP) and diffuse intrinsic pontine glioma (DIPG), and therefore presents an attractive target for therapeutic intervention. Through in silico virtual screenings and structure-activity relationship studies assisted by X-ray crystallographic analyses, a novel series of bis-heteroaryl pyrazole was identified as potent inhibitors of ALK2 (R206H). Derived from in silico hit compound RK-59638 (6a), compound 18p was identified as a potent inhibitor of ALK2 (R206H) with good aqueous solubility, liver microsomal stability, and oral bioavailability.
Organizational Affiliation:
Drug Discovery Chemistry Platform Unit, RIKEN Center for Sustainable Resource Science.